Nygen Analytics Announces the Release of CyteType
A Multi-Agentic Workflow for
Deep Cell Characterization
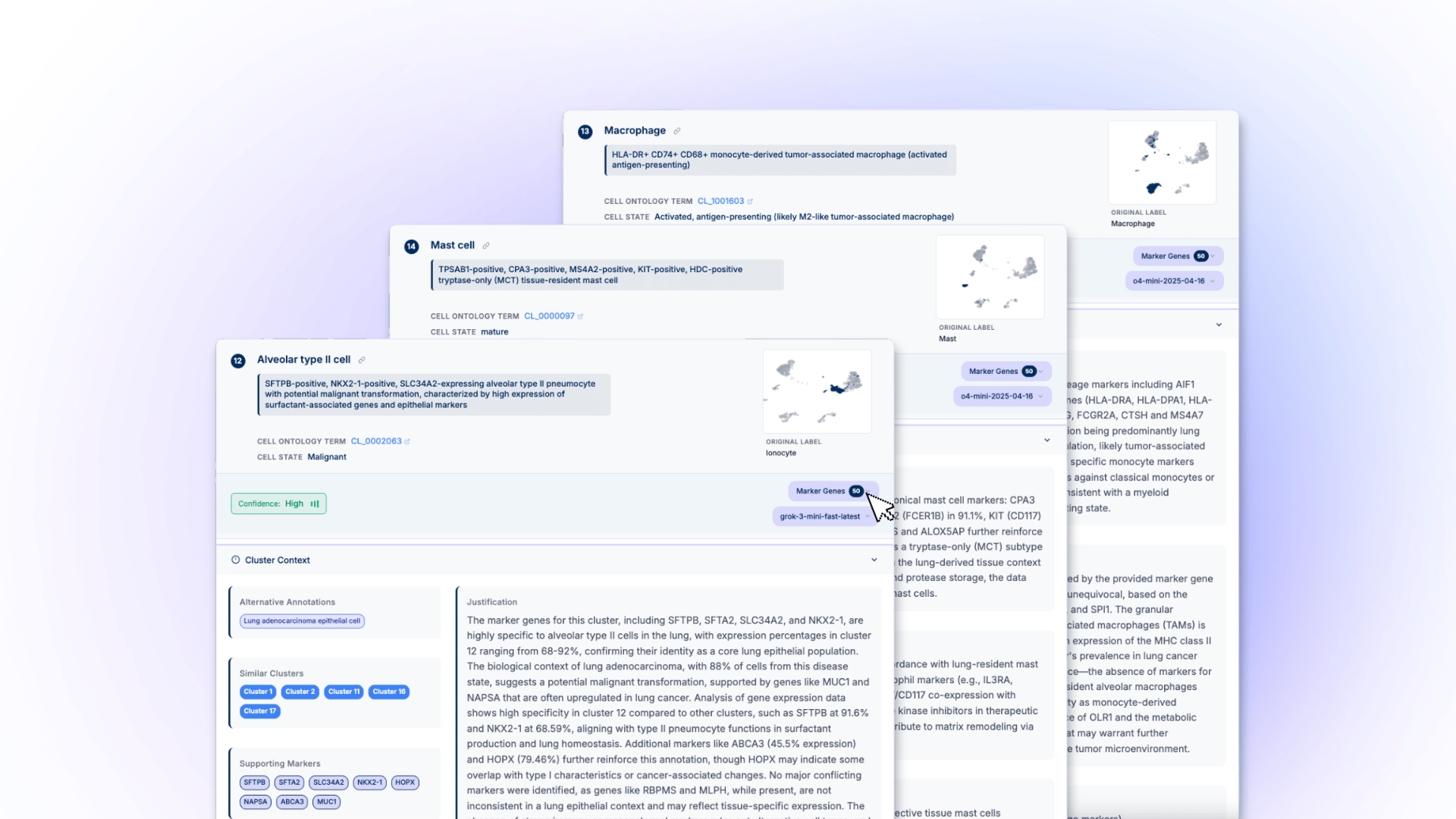
CyteType is designed to address the annotation bottleneck in single-cell research, setting a new standard in cell annotation and accelerating drug discovery.
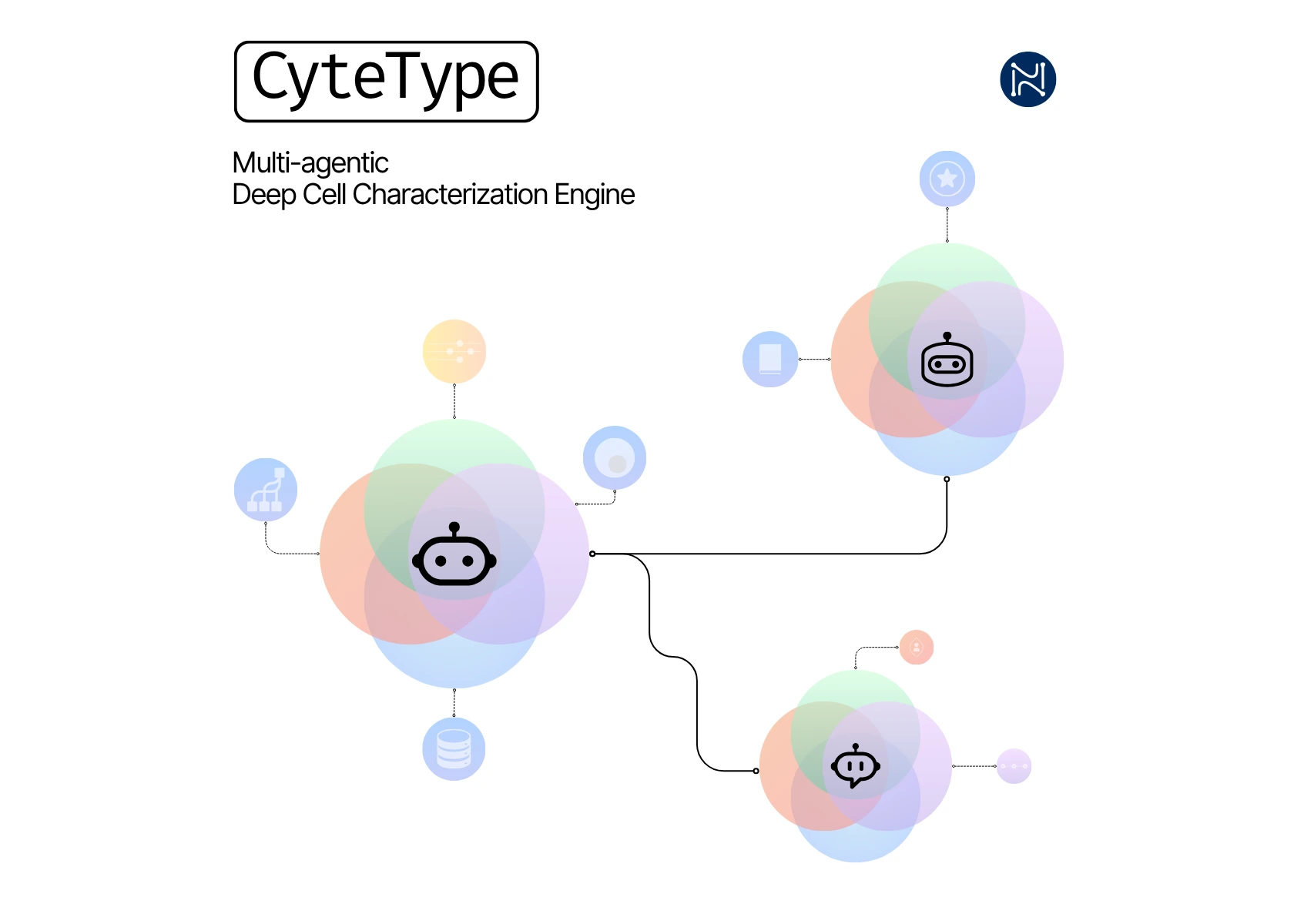
Malmö, Sweden | 23.07.2025 - Nygen Analytics has launched CyteType, a groundbreaking multi-agent AI platform that fundamentally reimagines how scientists approach single-cell annotation. Moving beyond simple pattern matching, CyteType employs three specialized AI agents that work collaboratively to deliver scientific reasoning, evidence validation, and contextual interpretation, mirroring how expert biologists actually think through complex annotation challenges.
At the core of CyteType's innovation is its revolutionary three-agent architecture: the Annotator dynamically analyzes entire transcriptomes and generates evidence-based hypotheses; the Reviewer acts as an independent validator, querying multiple biological databases and rejecting insufficient analyses; and the Corroborator provides literature integration with traceable citations. This collaborative approach ensures that every annotation is not just accurate, but scientifically defensible.
"Imagine hitting a key and instantly deploying a coordinated squad of virtual annotators, scientists, and project managers - all working together to review your single-cell data and assign accurate cell-type labels to each cluster. This is what CyteType is about. I like imagining these agents getting upset at each other, defending their own hypotheses while running bioinformatics scripts to prove their points, until they reach out a conclusion," Giovanni Marco Dall'Olio, Computational Biologist and early reviewer of CyteType.
Pathway-Level Reasoning Drives Deeper Insights
Traditional annotation tools rely on predetermined marker genes, often missing nuanced cell states that pure marker-based approaches overlook. CyteType's Annotator Agent queries expression levels of any gene in real-time, generating multiple hypotheses per cluster and ranking them by evidence strength.
- A standard tool might output:
- Cluster 1: T cells
- Top markers: CD3, CD4, CD8
- CyteType provides instead:
- CD4+ regulatory T cells (suppressive phenotype): High confidence annotation based on FOXP3, CTLA4, and IKZF2 expression. Alternative interpretations (early activation signature) considered and ruled out based on pathway analysis. Literature evidence supports tissue-resident Treg population with immunosuppressive function.
Built for Real Science
CyteType addresses critical gaps that have plagued AI biology tools:
- Evidence Tracking: Every claim is traceable to specific sources
- Uncertainty Quantification: Confidence scores reflect actual evidence strength
- Consistancy: Identical inputs produce identical outputs
- Interpretability: Full reasoning chains are available for inspection
The system processes up to 100+ concurrent clusters, with an average processing time of 2 minutes per cluster, and has been tested on datasets containing millions of cells. Early benchmarking shows significant improvements in identifying granular cell states that traditional methods collapse into broad categories.
Enterprise-Ready Architecture
CyteType's modular design integrates seamlessly with existing Python workflows through a simple pip install. The platform supports both cloud deployment and air-gapped enterprise environments, with options for AWS Bedrock or local model hosting.
Key features for research teams:
- Scalability: Serverless backend with intelligent rate limiting
- Security: Enterprise deployment options with complete data control
- Integration: Works directly with AnnData objects and existing workflows
- Flexibility: Bring your own LLM keys or use managed infrastructure
Community-Driven Innovation
CyteType is available now as an open-source platform. Nygen invites the scientific community to push the system to its limits; finding edge cases, challenging annotations, and helping build tools that don't just automate cell annotation, but make it better.
The real test isn't just performance metrics—it's whether CyteType can identify biologically meaningful cell states that other methods miss. Early results suggest it consistently discovers novel cell populations and disease-associated states that traditional approaches overlook entirely.
About Nygen Analytics
Nygen Analytics develops AI-native platforms for interpreting single-cell and spatial omics data. Combining domain-specialized LLMs, efficient data structures, and a deep understanding of translational biology, Nygen supports biopharma and academic researchers in uncovering actionable insights from high-dimensional data.
Press Contact
sukhitha.basnayake@nygen.io
www.nygen.io