Why We Built AI Agents That Actually Understand Single Cell Data
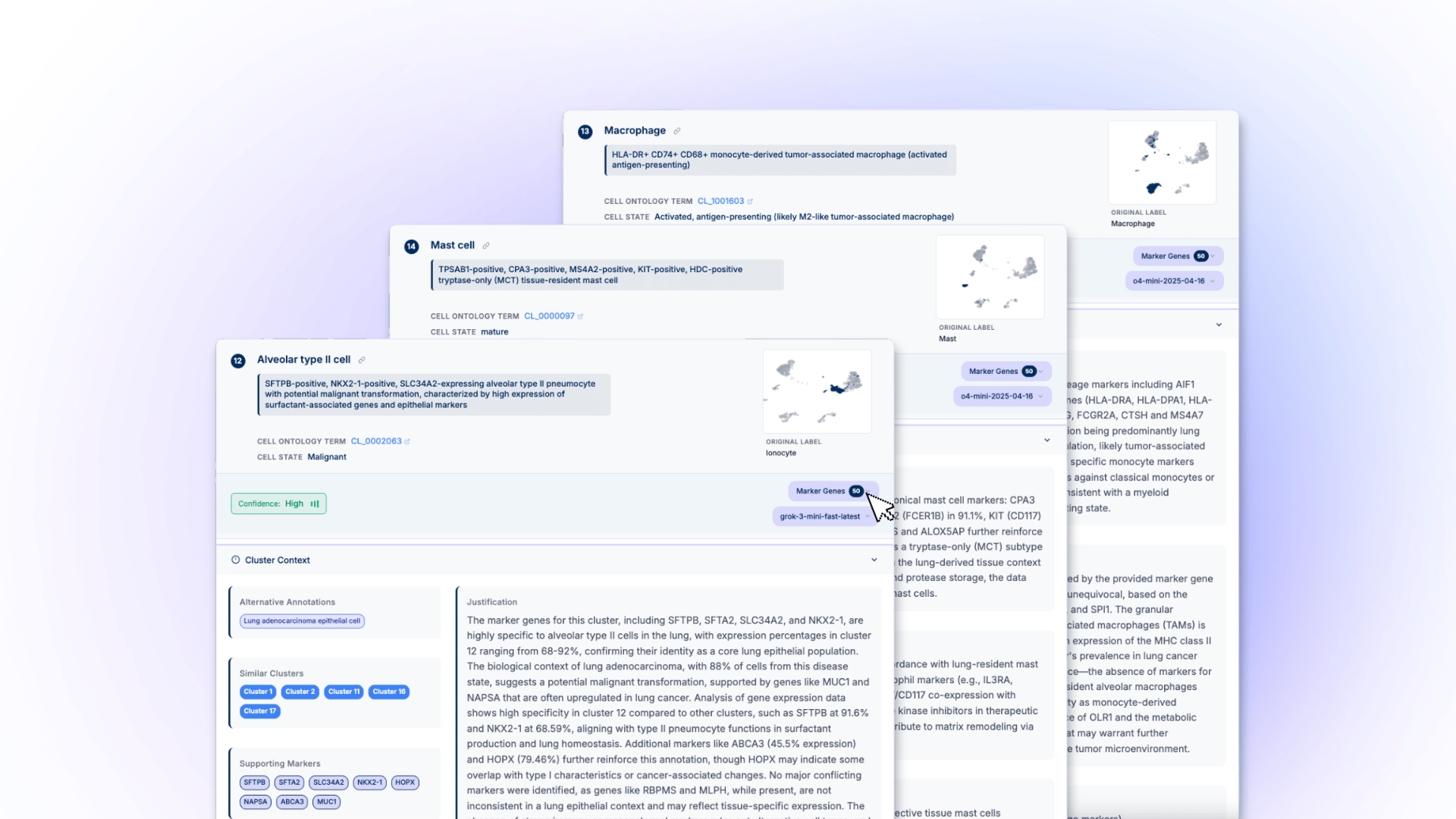
Breaking the Annotation Bottleneck for scRNA-seq Data Derived Cell Clusters
For over a year at Nygen, we've been obsessed with a deceptively simple question: Can large language models actually understand biology well enough to annotate single cell RNA-seq derived clusters?
The short answer is not in the way most people have been trying.
While others were building basic LLM wrappers around marker gene lists, we recognized that single-cell annotation requires scientific reasoning. It demands systematic evidence collection, contextual awareness, uncertainty quantification, and the ability to reconcile conflicting biological signals. That requires a team of specialized agents working in concert.
The Problem with "LLM's + Biology"
Most early attempts at LLM-based cell annotation follow a predictable pattern:
- Feed the model some marker genes
- Apply a reference atlast and "what cell type is this?"
- Hope for the best
This approach fails for the same reason asking a medical student to diagnose a patient based solely on symptoms fails. It lacks systematic evidence collection, context awareness, and the ability to reconcile conflicting information.
Traditional reference-based methods like Azimuth and SingleR are constrained by their training data and struggle with novel cell states, disease contexts, or species variations. They often collapse biologically distinct populations into broad categories, losing the cellular granularity that makes single-cell data valuable.
We needed to replicate how expert biologists actually think through complex datasets. not just automate existing approaches, but fundamentally improve the reasoning process itself.
How Our Agentic System Works
CyteType operates as a coordinated team of four specialized AI agents, each designed to spin up multiple sub-agents for rapid, deep annotation at scale. This distributed architecture enables parallel processing while maintaining biological coherence across datasets.
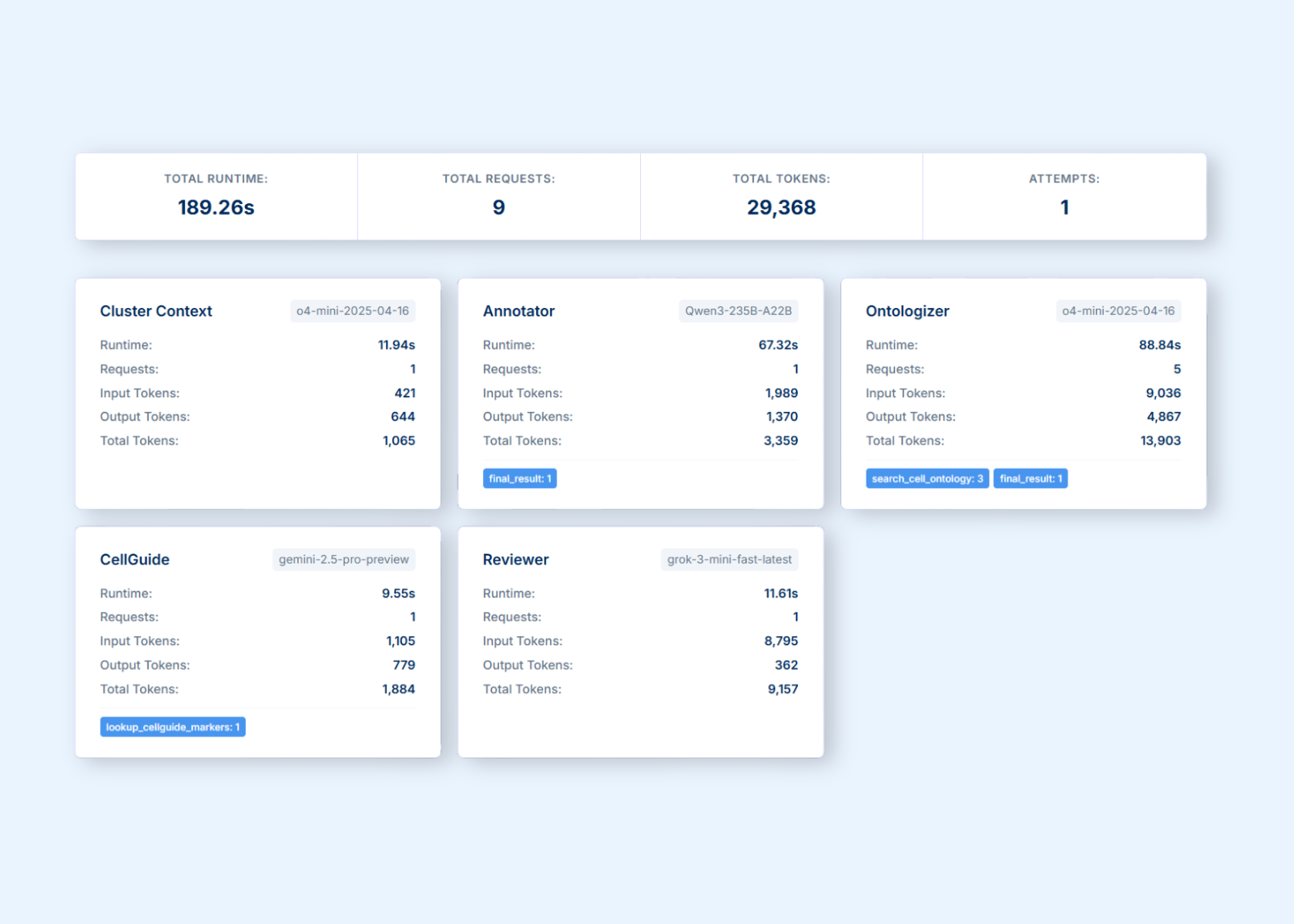
The Contextualization Agent
Builds comprehensive study context by synthesizing experimental metadata, treatment conditions, and sample information directly from AnnData objects. Sub-agents perform real-time queries to resources like GTEx to distinguish genuine biological signals from technical artifacts. The system updates context dynamically; insights from one cluster inform interpretation of related cell populations throughout the dataset.
The Annotation Agent
Orchestrates sub-agents to analyze complete expression profiles in real-time, querying any gene's expression level across clusters. For each cluster, coordinated sub-agents generate five ranked hypotheses based on transcriptomic evidence, pathway activity, and biological context. Specialized sub-agents detect interferon responses, cell cycle activity, and stress signatures that marker-based approaches miss.
The Reviewer Agent
Deploys validation sub-agents that independently collect evidence from Gene Ontology, GTEx, canonical marker databases, and PubMed Central. The reviewer can reject hypotheses and force re-analysis when evidence is insufficient, creating a feedback loop that maintains scientific rigor across the distributed system.
The Corroborator Agent
Orchestrates literature-focused sub-agents that identify experimental evidence, functional studies, and disease associations. Every annotation comes with traceable citations, ontology mappings, and confidence assessments coordinated through the agent's sub-agent network.
Real-World Validation Through Public Atlas Reannotation
We tested CyteType on the CELLXGENE multi-tissue atlas (ID: a3ffde6c-7ad2-498a-903c-d58e732f7470), targeting annotations that appeared biologically inconsistent.
What We Found
- Hillock/Club airway → luminal prostate epithelium
- Clusters 7 & 15 were luminal prostate epithelial cells, not "Hillock" or "Club" airway cells. This misclassification affected over 40,000 cells.
- Endothelial (lymphatic) → vascular smooth-muscle cell
- Putative lymphatic endothelium was re-labeled as vascular smooth-muscle cells based on comprehensive marker analysis.
- Ciliated epithelium → cycling epithelial progenitors
- Actively cycling cells mis-tagged as ciliated epithelium. These cells lacked FOXJ1 yet showed high NEK10/NEK11 expression.
- Endothelial (cardiac microvascular) → cardiac fibroblast
- Cardiac microvascular "endothelium" proved to be POSTN+/EMCN+ quiescent fibroblasts.
- Interstitial cells of Cajal → enteric neurons
- Cluster 28 was identified as enteric neurons, not interstitial cells of Cajal. PRKG1 (97%), DPP10 (90%), ANO1 (88%) override KIT contamination.
- "Unknown" clusters → keratinocytes
- Unknown clusters 73-74 revealed as keratinocytes through strong KRT1, KRT10 and involucrin expression.
Interactive Annotation Exploration
CyteType's report includes a chat interface for each annotated cluster, allowing researchers to probe the reasoning behind annotations. For clusters 7 & 15, we queried the system about the luminal prostate epithelium identification.
Query: "What specific markers distinguish these cells as luminal prostate epithelium rather than airway Hillock/Club cells?"
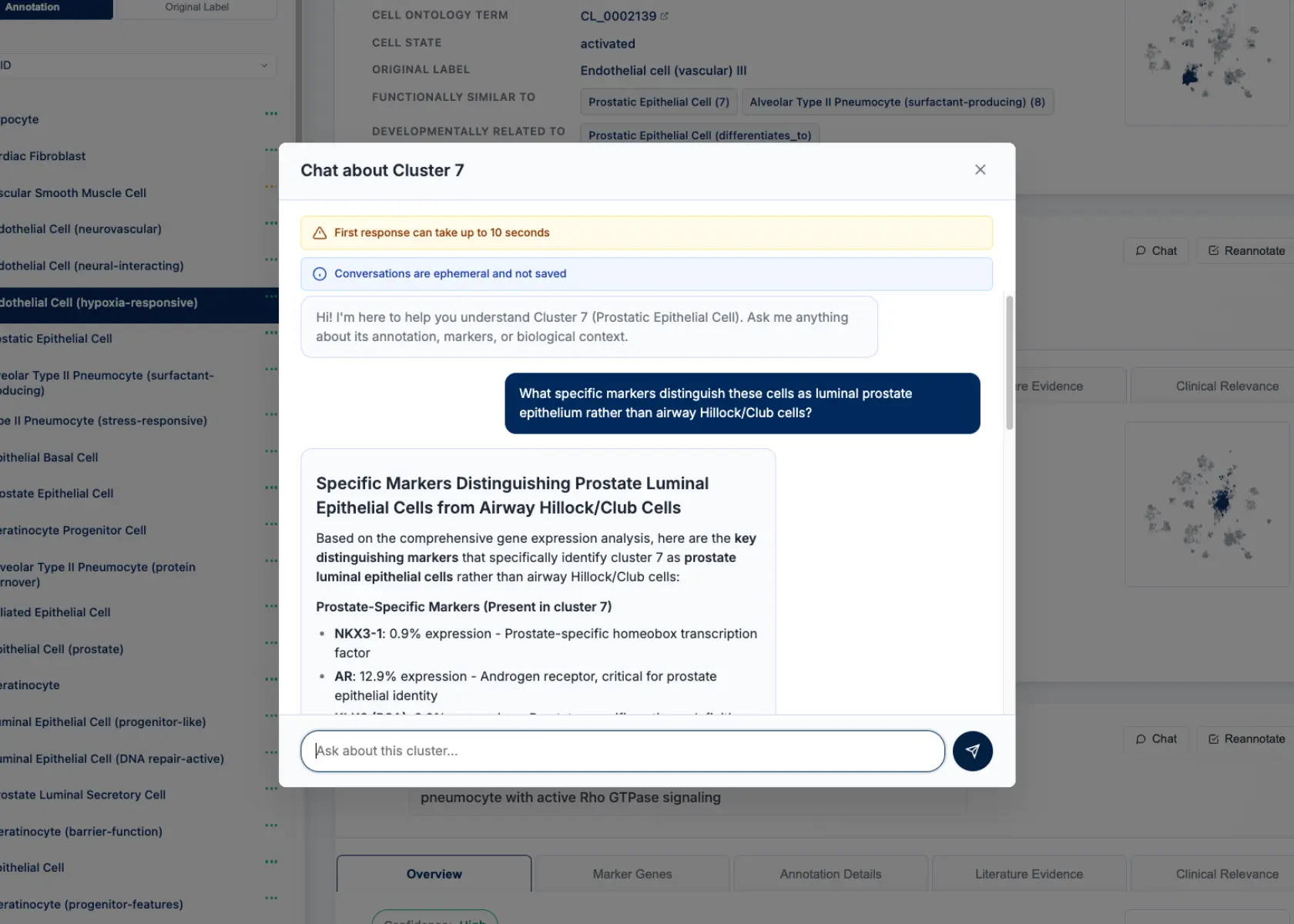
CyteType Response: "These clusters show strong expression of prostate-specific markers including KLK3 (PSA) at 89% expression frequency, NKX3-1 at 76%, and AR (androgen receptor) at 82%. Crucially, they lack canonical airway markers - SCGB1A1 (Club cell marker) shows only 3% expression, and FOXJ1 (ciliated cell marker) is absent. The transcriptomic profile aligns with luminal prostate epithelium from multiple prostate atlas references, not respiratory epithelium."
Users can also re-trigger annotation for specific clusters after providing additional biological context, enabling iterative refinement based on experimental knowledge or literature updates.
Why This Matters
CyteType's corrections shift pathway enrichment, cell-cell interaction maps and disease associations. The report shows that over 40,000 cells now align with prostate biology rather than airway, and fibroblast signals replace vascular signatures in cardiac tissue. These changes cascade into every model or hypothesis built on the atlas.
You can explore the dataset here.
Performance That Scales with Biology
CyteType processes clusters at 2 minutes each with 10 concurrent analyses, scaling to handle 100+ clusters simultaneously. CyteType consistently identifies granular cell states that other methods collapse into broad categories.
Instead of generic "T cells," CyteType delivers:
- CD4+ naive T cells (quiescent state)
- CD4+ T cells (early activation signature)
- CD4+ Th1 cells (IFN-γ high)
- CD4+ regulatory T cells (suppressive phenotype)
- CD4+ T cells (stress response active)
Each annotation includes specific pathway evidence, supporting literature, and confidence assessments, the detailed biological insight that drives meaningful research decisions.
The Human-AI Collaboration Loop
CyteType includes a feedback system where scientists can correct annotations directly in HTML reports. However, the Reviewer Agent independently evaluates user feedback, flagging suggestions that contradict biological evidence. This prevents confirmation bias while incorporating genuine domain expertise.
Infrastructure Built for Real Science
CyteType was designed from the ground up for scientific reasoning, not as a wrapper around existing models. The platform processes clusters at 2 minutes each with 10 concurrent analyses, scaling to handle 100+ clusters simultaneously while maintaining biological accuracy.
Every annotation links to specific literature, databases, and reasoning chains. Confidence scores reflect actual evidence strength rather than algorithmic certainty. Identical inputs produce identical outputs with full audit trails.
The platform integrates seamlessly with existing AnnData workflows through a simple Python client. For deployment, choose between our managed infrastructure or bring your own LLM keys. Enterprise users can deploy on AWS Bedrock or run completely air-gapped with local Ollama models. The serverless backend scales automatically with intelligent rate limiting to handle datasets of any size.
Full reasoning chains remain available for inspection, ensuring every biological claim can be traced back to its evidence sources.
Current Status and Development Roadmap
We're currently running comprehensive benchmarks against established methods like SingleR and CellTypist. Early results are promising, but we'll let the data speak for itself.
More importantly, we're seeing CyteType identify novel cell states and disease-associated populations that other methods miss entirely. That's where the real scientific value lies, not just automating what we already know, but discovering what we don't.
Try CyteType Yourself
import anndata
import scanpy as sc
import cytetype
# Load and preprocess your data
adata = anndata.read_h5ad("path/to/your/data.h5ad")
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
sc.pp.pca(adata)
sc.pp.neighbors(adata)
sc.tl.leiden(adata, key_added = "clusters")
sc.tl.rank_genes_groups(adata, groupby='clusters', method='t-test')
# Initialize CyteType (performs data preparation)
annotator = cytetype.CyteType(adata, group_key='clusters')
# Run annotation
adata = annotator.run(
study_context="Human brain tissue from Alzheimer's disease patients"
)
# View results
print(adata.obs.cytetype_annotation_clusters)
print(adata.obs.cytetype_cellOntologyTerm_clusters)CyteType is available now through pip install. The API is free to start, with deployment options from cloud infrastructure to air-gapped enterprise environments. Full documentation and examples are available on our GitHub repository.
We welcome all experiemntal researchers to test CyteType's limits. Find the edge cases. Challenge the annotations. Help us build annotation tools that advance biological understanding rather than simply automate existing practices..
Ready to dive in? Check out our documentation and start annotating with just a few lines of code. The future of single cell annotation is agentic.

