Multi-Agent AI for
scRNA-Seq Cell Type Annotation and Deep Characterisation
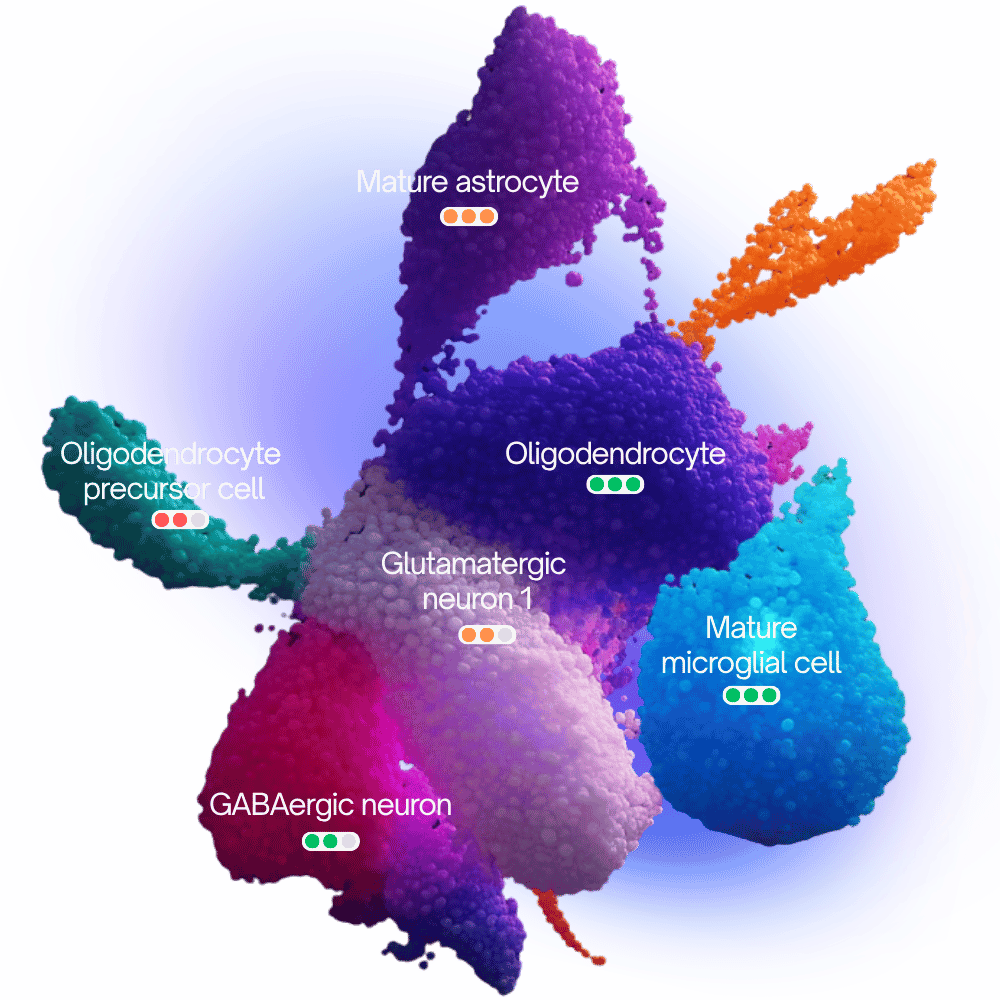
Stop settling for cell type labels. CyteType delivers mechanistic context explaining not just what cells are, but why they're in that state. Built for biopharma teams who need defensible annotations that accelerate target discovery.