CyteType: Multi-Agent AI for Single-Cell Annotation

The Annotation Bottleneck
Single-cell RNA sequencing datasets now routinely contain millions of cells. As computational methods have become increasingly efficient, the bottleneck has shifted from data analysis to biological interpretation, particularly in identifying cell type identities of clusters.
The numbers tell the story: reference-based classifiers trained on healthy tissue exhibit an accuracy drop of 15-30% when applied to disease samples. They miss rare cell types in approximately 20% of cases. Manual annotation can produce superior results, but it's time-consuming and shows 25% inter-annotator variability. Current methods provide only cell type labels without justification or caveats, limiting result interpretability.
Large language models have shown potential. GPT-4 achieved 75% agreement with expert annotations in published assessments. But existing LLM approaches have significant limitations: they process only top marker genes rather than full expression profiles, rely on static knowledge from pretraining data, and lack mechanisms to validate predictions against external databases or quantify uncertainty.
The Current Landscape
Cell type annotation methods fall into several categories, each with distinct trade-offs.
Reference-based methods like SingleR and CellTypist compare query cells against pre-annotated reference datasets. They work well when query data closely matches training distributions but degrade under domain shift. When applied to disease contexts, tumour microenvironments, or tissues not well-represented in training data, accuracy suffers substantially.
Marker-based approaches such as Garnett and scType use curated marker gene lists. These require extensive manual curation and struggle with cell types that share overlapping marker profiles. They also cannot easily adapt to novel or transitional cell states.
Transformer-based foundation models like scGPT, GeneFormer, and TranscriptFormer learn representations from large-scale single-cell atlases. They show promise for transfer learning but require careful fine-tuning for novel contexts, and their predictions remain difficult to interpret biologically.
LLM-based methods like GPTCellType leverage the broad biological knowledge encoded in large language models. However, they typically process only top marker genes, lack external validation mechanisms, and provide no uncertainty quantification. The LLM's knowledge is frozen at training time, disconnected from current literature and databases.
Architecture Over Model Selection
We developed CyteType to address these limitations through a different approach: rather than switching to a more powerful model, we designed a multi-agent architecture that structures the annotation task itself.
CyteType is built around a five-agent framework, where each agent handles a distinct aspect of the scientific reasoning process.
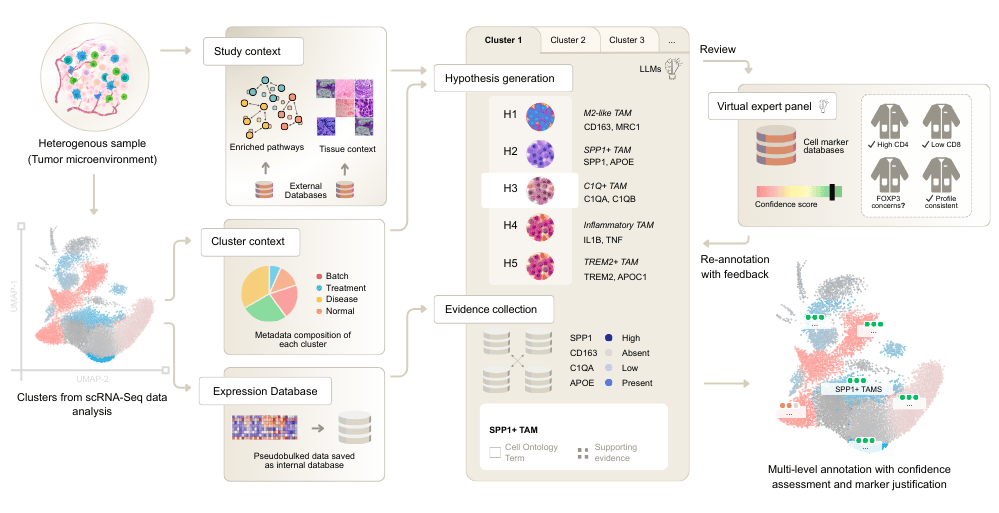
The Contextualizer agent establishes biological ground truth before annotation begins. It infers organism, tissue, and pathway context from the data and metadata, selecting appropriate pathway databases and generating cluster-specific context. This agent integrates with GTEx, Enrichr (Gene Ontology, Reactome, WikiPathways), and blitzGSEA.
The Annotator agent employs hypothesis-driven reasoning. Rather than producing a single prediction, it generates multiple competing cell type hypotheses and systematically tests each against the full expression profile. It collects evidence dynamically by querying a pseudobulked expression database, then selects the top hypothesis based on evidence strength and links it to Cell Ontology terms.
The Reviewer agent provides critical validation through a simulated expert panel. It assesses annotation confidence by checking predictions against external reference databases like CellGuide, detects cellular heterogeneity within clusters, and triggers re-annotation when needed. This creates an interpretable "trust layer" that existing methods lack.
The Literature & Clinical Relevance agent connects annotations to current knowledge. It searches PubMed for supporting literature, identifies disease associations via Disease Ontology, and flags potential drug targets through Drug Ontology integration.
The Summarizer agent synthesizes results across the entire study. It performs annotation similarity analysis, disambiguates naming inconsistencies, and generates semantic cluster ordering to produce a coherent biological narrative.
Benchmarking: Architecture vs. Model
Rigorous benchmarking of cell type prediction algorithms has proven challenging due to the free-text nature of cell type labels. To address this, we developed CyteOnto, a semantic similarity framework that maps cell type labels to Cell Ontology terms using text embeddings. Unlike string-matching or graph-based methods that fail to capture semantic relationships, CyteOnto quantifies normalized distances between ontology terms to derive similarity scores ranging from 0 to 1.
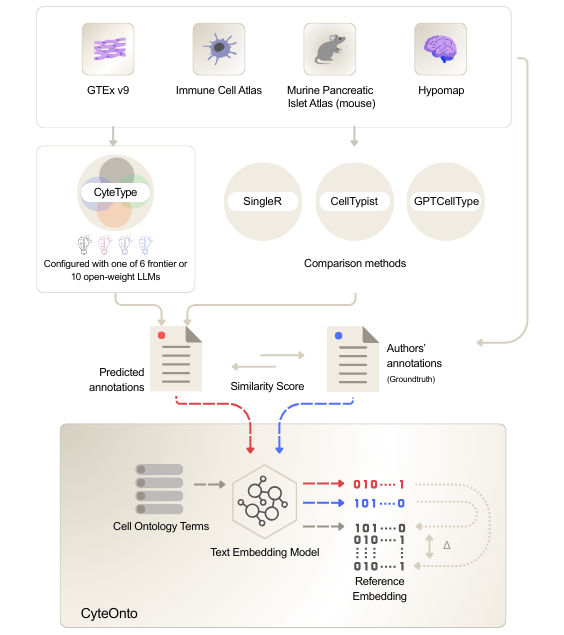
We evaluated CyteType on four benchmark datasets spanning 205 clusters across diverse biological contexts: HypoMap (developing mouse brain), Immune Cell Atlas (human immune cells), GTEx v9 (cross-tissue human atlas), and Mouse Pancreatic Cell Atlas.
To isolate architectural benefits from underlying LLM capabilities, we compared CyteType against GPTCellType using identical models (GPT-5), alongside established reference-based methods CellTypist and SingleR.
The results were clear:
- CyteType vs. GPTCellType (same LLM): 388.52% higher similarity score (z = 8.04, p < .001, Tukey-adjusted)
- CyteType vs. CellTypist: 267.9% higher average similarity score (z = 8.15, p < .001)
- CyteType vs. SingleR: 100.67% higher average similarity score (z = 8.36, p < .001)
The comparison against GPTCellType is particularly informative. Both methods used GPT-5, yet CyteType's structured hypothesis testing and evidence collection dramatically outperformed direct LLM prompting. This demonstrates that architectural innovations provide value independent of the underlying language model.
Model Flexibility Without Sacrificing Performance
To assess robustness, we benchmarked 16 LLMs spanning closed-weight models (GPT-5, Claude Sonnet 4, Gemini 2.5 Pro) and open-weight models (DeepSeek R1, Kimi K2, Qwen3 Thinking).

Closed-weight models achieved the highest mean similarity scores. Open-weight models showed modestly decreased performance but still outperformed traditional methods substantially. Notably, LLMs with built-in chain-of-thought reasoning demonstrated no significant advantage over standard models (b = 0.014, SE = 0.011, p = 0.22), suggesting CyteType's structured workflow supersedes model-native reasoning capabilities.
This has practical implications. Researchers can select models based on cost and privacy requirements without sacrificing the benefits of CyteType's architecture. Closed-weight models maximize accuracy for critical applications. Open-weight models like Kimi K2 and DeepSeek R1 achieve 95% of peak performance at lower cost, suitable for exploratory analysis or budget-constrained settings.
More Than Just Label Assignment
CyteType's Reviewer agent generates calibrated confidence scores and heterogeneity flags that enable targeted expert review. High-confidence annotations were significantly more likely to have higher similarity scores than low-confidence ones (F = 23.88, p < .001). Heterogeneous clusters showed significantly lower similarity scores against author labels (F = 8.45, p < .01).
Across repeated runs, median majority agreement exceeded 80% for all LLMs, with consensus Cell Ontology labels achieved for more than 70% of clusters. Both reviewer confidence and heterogeneity flags predicted similarity scores even after accounting for LLM choice.
These metrics transform cell type annotation from black-box classification into a transparent process with quantified uncertainty.
Discovery, Not Just Classification
Applying CyteType to 977 clusters across 20 datasets revealed systematic patterns. Rather than simply validating existing annotations, CyteType added value across all scenarios:
- 41% of clusters received functional enhancement (adding cell state information)
- 29% underwent refinement to specific subtypes
- 30% required major reannotation
Annotations mapped to 327 unique Cell Ontology terms with high diversity (no term exceeding 2.5% frequency) and identified 116 distinct cell states, with activation (37%) and maturation (16%) most prevalent.
High-confidence discrepancies occurred predominantly in disease contexts where reference methods historically underperform. For example, in the diabetic kidney disease atlas, author-denominated parietal epithelial cells and leukocytes were relabelled as injured proximal tubule cells (ALDH1A2+, CFH+, VCAM1+) and T cells (activated CD45+DOCK2+ pro-inflammatory), respectively.
This distribution demonstrates that CyteType functions not merely as a classifier but as a discovery tool, uncovering biological context missed by reference-based methods.
Availability and Deployment
CyteType is available as open-source software under CC BY-NC-SA 4.0 license:
- Python (AnnData compatible): Install via pip (
pip install cytetype) from GitHub - R (Seurat compatible): CyteTypeR is available from GitHub
Both implementations generate comprehensive reports that integrate directly into existing workflows. Cell type annotations are returned into AnnData/Seurat objects along with Cell Ontology terms for downstream analysis. Interactive HTML reports provide detailed marker justification, pathway enrichment, literature evidence, and disease associations, with embedded chat functionality enabling iterative refinement.
For enterprise deployments requiring data sovereignty, CyteType supports locally hosted open-weight models via Ollama, enabling air-gapped operation without external API calls.
The Architecture Principle
CyteType addresses fundamental limitations in current annotation approaches by transforming cell type identification from classification into evidence-based characterization. While reference-based methods constrain predictions to previously characterized cell types and degrade under domain shift, and transformer-based foundation models require careful fine-tuning for novel contexts, CyteType's hypothesis-testing framework discovers cell states and rare populations with calibrated uncertainty.
Recent LLM approaches have demonstrated that generative models can produce interpretable annotations. However, our direct comparison using identical models shows that multi-agent architecture provides substantial value beyond the capabilities of any LLM alone.
LLMs are not magic. But well-architected agents that structure scientific reasoning, ground predictions in evidence, and quantify uncertainty? They can be transformative.
Ready to dive in? Check out our documentation and start annotating with just a few lines of code. The future of single cell annotation is agentic.

